Personal Details

I am finalizing my Ph.D. at the Quantitative Biology Center, Tübingen, with expertise in pangenomics and multiomics. I develop methods to analyze (pan)genomic variation, manage interdisciplinary projects, and create efficient tools like nf-core/pangenome for large-scale pangenomic analysis. I provide guidance to biomedical researchers on experimental design, data analysis, and multiomics integration. I also spearhead hackathons and workshops to foster innovation and knowledge-sharing within the bioinformatics community. Outside of work, I enjoy an active lifestyle and watching Sesame Street for a good laugh.
Chat | Publications | Code | ResearchGate | ORCID | Linkedin | X | 
Employment
1 May 2025 to …
Senior Bioinformatics Scientist at Illumina (remote).
Reporting to Dr. Konrad Scheffler.
-
DRAGEN C++ Developer
1 Jul. 2019 to 30 Jun. 2024 (5 years)
PhD student at the Quantitative Biology Center (QBiC) (Tübingen, Germany).
Evaluation: Summa cum laude.
Reporting to Prof. Sven Nahnsen.
-
Pangenome graph software & research:
-
Developed nf-core/pangenome, a cluster-efficient pipeline for constructing pangenome graphs without bias or exclusion, improving execution speed by 2-3 (Heumos et al. 2024, Bioinformatics) ( nf-core/pangenome)
-
Co-developed ODGI, the de-facto standard for pangenome graph analysis (Guarracino*, Heumos* et al. 2022, Bioinformatics) ( ODGI)
-
Designed and co-developed PG-SGD, a pangenome graph layout algorithm that achieved 10x faster performance and reduced memory usage by 2x, cutting layout calculation time from over a week to one day for human pangenomes (Heumos*, Guarracino* et al. 2024, Bioinformatics) ( PG-SGD)
-
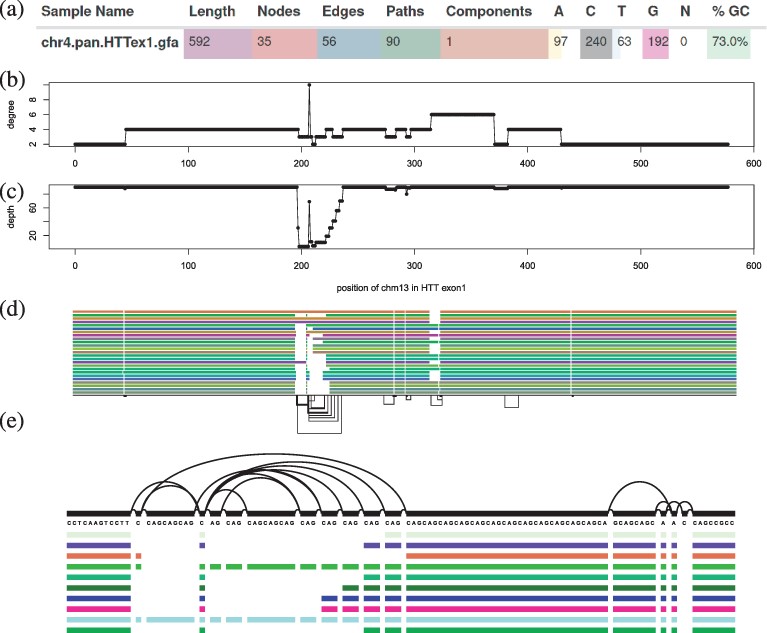
Major contributor to the PanGenome Graph Builder (Garrison*, Guarracino*, Heumos et al. 2024, Nature Methods) ( PGGB)
-
Evaluating pangenome graphs ( PGGE)
-
Supporting efforts for efficient pangenome graph indices (Balaz et al. 2024, LATIN2024: Theoretical Informatics)
-
Contributed to a Pangenome Graphs review (Eizenga et al. 2020, Annual Review of Genomics and Human Genetics)
-
As an associate member of the Human Pangenome Reference Consortium I contributed to building the first draft human pangenome reference (Liao*, Asri*, Ebler* et al. 2023, Nature)
-
Industry collaboration with Computomics GmbH on coding a pangenome browser Pantograph. Development of pangenome graph browser using React, MobX-State-Tree, JavaScript ( Genome Graph Browser).
-
Experimenting towards an interface between RDF/SPARQL and pangenome graphs together with Jerven Bollemann and Toshiyuki T. Yokoyama winning an ISMB 2020 Best Poster Award: Semantic Variation Graphs: Ontologies for Pangenome Graphs
-
-
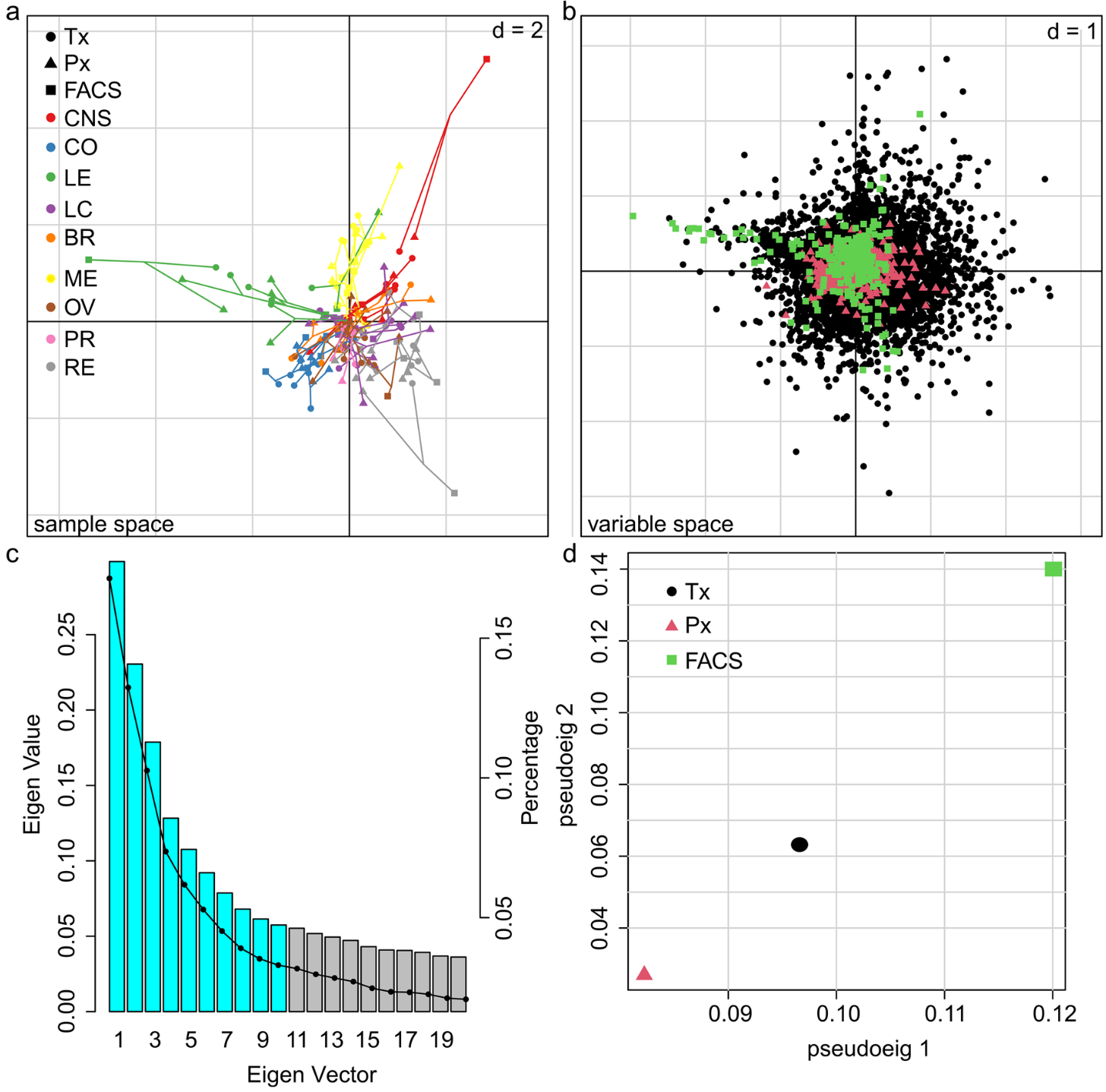
Multiomics analysis of the NCI-60 tumor cell panel (Heumos*, Dehn* et al. 2022, Cancer Cell International) ( QMSFC):
-
Crawling and integrating multiomics data from resources like TCGA
-
Differential expression analysis of RNA-Seq data
-
Integrative analysis of transcriptomic microarray data (Affymetrix)
-
Curation, quality control, differential expression analysis of Fluorescence Activated Cell Sorting (FACS) data
-
Proteomics and phosphoproteomics data curation, and differential expression analysis
-
Reverse Phase Protein Array (RPPA) differential expression analysis
-
-
Organizer, tutor, and chair of international hackathons and workshops
-
Managing virtual machines and users in QBiC’s deNBI cloud instances
-
Organization of retreats
-
Mentoring undergraduates
-
Learning ONT sequencing and base calling at PANGAIA’s Winter Wet Lab school
1 Jul. 2017 to 30 Jun. 2019 (2 years)
Research assistant at the Quantitative Biology Center (QBiC) (Tübingen, Germany).
Reporting to Dr. Stefan Czemmel
Employer’s reference
-
Member of bioinformatics support and project management team (BioPM)
-
Bridge function to the infrastructure and scientific software team
-
Counseling of biomedical researchers on experimental design, data analysis, and paper writing
-
FACS data analysis
-
Proteomics LFQ data analysis
-
Germline variants analyses on whole-genome sequencing (WGS) data: Quality control, read trimming and mapping, variant calling, and functional prediction
-
Compilation of highly standardized and reproducible bioinformatics pipelines
-
Excellent customer service when performing data processing and statistical analysis of big biomedical data
-
Initiation and maintenance of a QBiC report template for analysis results
-
Driving force behind standardized SOPs to improve QBiC’s infrastructure
-
Shaped research grant application at the Ministry for Economics and Energy (BMWi) titled PANTOGRAPH aquiring 190,000€ for researching pangenome graph visualization
-
Supervision of student projects
-
After 1 year: Vice coordinator of the BioPM team
1 Nov. 2016 to 31 May 2017 (7 months)
Master student at the Computomics GmbH (Tübingen, Germany)
Reporting to Björn Geigle and Dr. Jörg Hagmann
Thesis Interactive Visualization of Genome Variation Graphs
Evaluation: 1.0
ISMB 2017 Best Poster Prize Interactive pangenome visualization using variant graphs
Tooling: Interactive full-stack web application with Node.js, nbind to make C++ VG accessible in JavaScript, Pug, D3, HTML, CSS ( AG)
1 Jun. 2015 to 31 Sep. 2016 (1 year, 2 months)
Research student at the Max Planck Institute for the Science of Human History (Jena, Germany)
Reporting to Dr. Alexander Herbig in the Computational Pathogenomics research group
-
Development of bioinformatics programs for the analysis of paleogenetic NGS data ( GenConS)
( Fellows Yates et al. 2017, Scientific Reports)
1 Sep. 2014 to 31 May 2015 (11 months)
Research student at the Institute for Archaeological Sciences (Tübingen, Germany)
Reporting to Dr. Alexander Herbig in the Paleogenetics research group
-
Performance of system administration tasks and development of bioinformatics programs for the analysis of paleogenetic NGS data
1 Jul. 2014 to 30 Aug. 2014 (1year, 1 month)
Research student at the MFT Services (Tübingen, Germany)
Reporting to Dr. Günter Jäger in the Medical Genetics research group
-
Analysis of RNA-Seq data
1 Jun. 2013 to 31 Mar. 2014 (10 months)
Research student at the Centre for Bioinformatics (Tübingen, Germany)
Reporting to Prof. Kay Nieselt in the Integrative Transcriptomics research group
-
Analysis of RNA-Seq data and extension of an in-house developed Java tool for the analysis of RNA-Seq data
1 Aug. 2010 to 31 Aug. 2010 (1 month)
1 Aug. 2011 to 31 Aug. 2011 (1 month)
1 Aug. 2012 to 31 Aug. 2012 (1 month)
Temporary shipping assistant at WALDNER (Wangen, Germany)
-
Packaging small part materials and transporting them to the loading area
1 Jul. 2009 to 31 Mar. 2010 (9 months)
Alternative service as an ambulance man at the DRK Rettungsdienst Bodensee-Oberschwaben gGmbH (Ravensburg, Germany)
-
Operations took place both in the ambulance and in the patient transport vehicle
1 Feb. 2007 to 31 Jul. 2010 (3 years, 5 months)
Side job at the PEKANA Naturheilmittel GmbH (Kisslegg, Germany)
-
Assistant for office, shipping, and pharmaceutical packaging tasks
Education
1 Jun. 2019 to 28 Mar. 2025
PhD student at the Quantitative Biology Center (QBiC) (Tübingen, Germany).
Reporting to Prof. Sven Nahnsen.
-
Thesis Computational methods for pangenomics and multiomics integration
1 May 2014 to 30 May 2017
MSc Bioinformatics at the University of Tübingen (Tübingen, Germany).
MSc certificate
-
Major fields:
-
NGS
-
RNA Biology
-
Java + JavaFX development
-
Data management in quantitative biology
-
-
Thesis Interactive Visualization of Genome Variation Graphs at Computics GmbH: 1.0
1 Oct. 2010 to 30 Apr. 2014
BSc Bioinformatics at the University of Tübingen (Tübingen, Germany).
BSc certificate
-
Major fields:
-
Functional programming with Scheme
-
Basics Bioinformatics
-
Java + JavaSwing development
-
Linux
-
-
Thesis TOPAS - TOolkit for Processing and Annotating Sequence data: 1.3
1 Aug. 2000 to 30 Jun. 2009
Abitur (Wangen, Germany).
School certificate
Publications
*first authorship
| Journal | Title | Contribution | Links |
|---|---|---|---|
Bioinformatics, 2024 |
Cluster-efficient pangenome graph construction with nf-core/pangenome |
Pipeline conception, software development, testing, documentation, design and conduction of experiments, paper writing |
|
SC24: International Conference for High Performance Computing, Networking, Storage and Analysis, 2024 |
Rapid GPU-Based Pangenome Graph Layout |
Guidance on algorithm implementation, feedback of the cache optimized CPU and GPU implementations, read & criticized & edited manuscript |
|
PLOS Computational Biology, 2024 |
nf-core/airrflow: an adaptive immune receptor repertoire analysis workflow employing the Immcantation framework |
Software development, paper editing |
|
Nature Methods, 2024 |
Building pangenome graphs |
Software development, documentation, testing, contributed to Figure 1, wrote Section A1, made Figure A1, and contributed to paper writing and editing |
|
Bioinformatics, 2024 |
*Pangenome graph layout by Path-Guided Stochastic Gradient Descent |
Algorithm implementation leader, testing, documentation, design and conduction of experiments, paper writing |
|
LATIN 2024: Theoretical Informatics, 2024 |
Wheeler Maps |
Advisor for the integration of a wheeler maps implementation with real life pangenome graphs, built and provided initial pangenome graphs for testing the implementation, manuscript editing |
|
Acta Neuropathologica, 2023 |
Integrative proteomics highlight presynaptic alterations and c-Jun misactivation as convergent pathomechanisms in ALS |
Paper editing, LFQ proteomics analysis |
|
Nature, 2023 |
A draft human pangenome reference |
Paper editing, pangenome graph creation and visualization |
|
Cancer Cell International, 2022 |
*Multiomics surface receptor profiling of the NCI-60 tumor cell panel uncovers novel theranostics for cancer immunotherapy |
Data curation and quality control, performed the MCIA, RNAseq analysis and TCPA data exploration, wrote methods sections of the software tools and steps I applied, generated visualizations for Figures 1-3, and manuscript editing |
|
Bioinformatics, 2022 |
*ODGI: understanding pangenome graphs |
Paper and documentation writing, performance evaluation, testing, implemented several tools |
|
Frontiers in Plant Science, 2022 |
A Perspective on Plant Phenomics: Coupling Deep Learning and Near-Infrared Spectroscopy |
Experimental counseling, data management |
|
Frontiers in Immunology, 2020 |
Specific Induction of Double Negative B Cells During Protective and Pathogenic Immune Responses |
Data curation |
|
Bioinformatics, 2020 |
Efficient dynamic variation graphs |
Implementation of some ODGI subcommands (pathindex, server, panpos), optimization of one (bin), documentation writing for ODGI |
|
Annual Review of Genomics and Human Genetics, 2020 |
Pangenome Graphs |
I made Table 1 and contributed to Sections 4.4 and 6.1 and Figure 2, paper editing |
|
Frontiers in Immunology, 2019 |
Platelets Aggregate With Neutrophils and Promote Skin Pathology in Psoriasis |
FACS data analysis |
|
Frontiers in Immunology, 2019 |
PSM Peptides From Community-Associated Methicillin-Resistant Staphylococcus aureus Impair the Adaptive Immune Response via Modulation of Dendritic Cell Subsets in vivo |
Statistical analysis counseling, paper editing |
|
Nature Scientific Reports, 2017 |
Central European Woolly Mammoth Population Dynamics: Insights from Late Pleistocene Mitochondrial Genomes |
GenConS software development and testing, wrote section about GenConS, paper editing |
Talks & Posters
| Time | Conference | Title | Links |
|---|---|---|---|
|
Cluster efficient pangenome graph construction with nf-core/pangenome |
||
|
HPRC HUGO24 Workshop Rome |
Building and Analyzing Pangenome Graphs |
|
|
Cluster efficient pangenome graph construction with nf-core/pangenome |
||
|
nf-core community, virtual |
Cluster scalable pangenome graph construction with nf-core/pangenome |
|
|
Cluster scalable pangenome graph construction with nf-core/pangenome |
||
|
Pangenome Graphs |
||
|
Pangenome Graphs |
||
|
Graph layout by path-guided stochastic gradient descent |
Talk |
|
|
Graph Layout by Path-Guided Stochastic Gradient Descent |
||
|
Exploring pangenome graphs and possible applications |
||
|
A pangenome for the expanded BXD family of mice |
||
|
ODGI: scalable tools for pangenome graphs |
||
|
The PanGenome Graph Builder |
||
|
The PanGenome Graph Builder |
||
|
Graph Layout by Path-Guided Stochastic Gradient |
||
|
Graph Layout by Path-Guided Stochastic Gradient |
||
|
Pantograph: Scalable Interactive Graph Genome Visualization |
||
|
Semantic Variation Graphs - A Pangenome Ontology |
||
|
Semantic Genome Graphs |
||
|
VG Browser: Interactive Visualization of Genome Variation Graphs |
Awards
| Time | Place | Description | Links |
|---|---|---|---|
|
Student Travel Award 700 CHF (720.32 EUR) |
||
|
Best Poster Award Semantic Variation Graphs: Ontologies for Pangenome Graphs |
||
|
Best Poster Award Interactive pangenome visualization of variant graphs |
||
|
Bronze Award |
Grants
| Time | Sponsor | Amount | Description |
|---|---|---|---|
|
190,000 EUR |
Research grant Pantograph together with Computomics GmbH to research pangenome graph visualization. |
Invitations
| Time | Place | Description | Links |
|---|---|---|---|
|
MemPanG24 Pangenomics, University of Tennessee Health and Science Center, Memphis, USA |
Invited Organizer, instructor, and chair |
|
|
HPRC Pangenomics Workshop at HUGO 2024, Aula Multimediale Rettorato, Sapienza University of Rome, Rome, Italy |
Invited Instructor |
|
|
MemPanG23 Pangenomics, University of Tennessee Health and Science Center, Memphis, USA |
Invited Organizer, instructor, and chair |
|
|
nf-core community, virtual |
Invited talk nf-core bytesize talks 2023 Cluster scalable pangenome graph construction with nf-core/pangenome |
|
|
Invited talk Exploring pangenome graphs and possible applications |
Hackathons
| Time | Info | What | Links |
|---|---|---|---|
|
nf-core hackathon, virtual |
Co-team leader group pipelines, finalizing nf-core/pangenome |
|
|
nf-core hackathon, Barcelona, Spain |
Progressing nf-core/pangenome |
|
|
nf-core hackathon, virtual |
Progressing nf-core/pangenome |
|
|
nf-core hackathon, virtual |
Progressing nf-core/pangenome |
|
|
Pangenomics BioHacking, Online, Virtual in Milano |
Expert for pangenome graph construction and participant |
|
|
ELIXIR Europe Biohackathon, Barcelona, Spain |
Progressing nf-core/pangenome |
|
|
nf-core hackathon, virtual |
Starting nf-core/pangenome |
|
|
ELIXIR Europe Biohackathon, virtual |
Project leader Federated Interoperable Annotated Variation Graphs |
|
|
Crusco Biohackathon, Lavello, Italy |
Progressing PG-SGD with Andrea Guarracino and Erik Garrison |
|
|
COVID-19 Biohackathon, virtual |
Co-Project leader Pangenome Browser and co-project leader Pangenome Ontology |
COVID-19 Biohackathon 2020 |
|
Computomics GmbH, Tübingen, Germany |
Progressing Pantograph, playing around with pangenome graphs and SPARQL |
|
|
NBDC DBCLS BioHackathon 2019, Fukuoka, Japan |
Co-Project leader Pantograph, playing around with SequenceTubeMap and SPARQL |
Hackathon Wiki |
Programming Languages
| Start | Language | Projects |
|---|---|---|
|
Scheme |
Learning to code during my first three semesters BSc Bioinformatics |
|
Java |
BSc thesis TOPAS - TOolkit for Processing and Annotating Sequence data ( TOPAS) |
|
Bash |
Contributing to the PanGenome Graph Builder ( PGGB). Implementing the PanGenome Graph Evaluation tool ( PGGE). |
|
R |
Performing multiomics analysis ( QMSFC). |
|
Python |
Implementing an ODGI MultiQC module ( ODGI MultiQC Module). Supervision of graph genome browser implementation ( Pantograph). |
|
Javascript |
Supervision of graph genome browser implementation ( Pantograph). Interactive full-stack web application with Node.js, nbind to make C++ VG accessible in JavaScript, Pug, D3, HTML, CSS ( AG) |
|
C++ |
|
|
Nextflow |
Sole developer of nf-core/pangenome. |
|
Rust |
GAF alignment evaluation tool ( rs-peanut) |
Teaching
| Time | Course | Role | Links |
|---|---|---|---|
|
MemPanG24 Pangenomics, University of Tennessee Health and Science Center, Memphis, USA |
Organizer, instructor, and chair, created new material and tutorials, held lessons, assisted the participants, tested the VMs |
|
|
HPRC Pangenomics Workshop at HUGO 2024, Aula Multimediale Rettorato, Sapienza University of Rome, Rome, Italy |
Instructor, updated material and tutorials, held lessons, assisted the participants |
|
|
M3 Pangenome Workshop, M3 Research Center, University of Tübingen, Tübingen, Germany |
Speaker, live demonstration of nf-core/pangenome |
|
|
MemPanG23 Pangenomics, University of Tennessee Health and Science Center, Memphis, USA |
Organizer, instructor, and chair, created new material and tutorials, held lessons, assisted the participants |
|
|
Biomedical Data Management, University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
Tutor, managed seminar, hold tutorials, exam assistant |
NA |
|
Grundlagen der Bioinformatik, University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
Tutor, hold tutorials, revied and updated practical lessons, exam grader |
|
|
Advanced Bioinformatics: Data Mining and Data Integration for Life Science (1.5 CFU/ECTS), Master’s degree, Utrecht Bioinformatics Center, Utrecht, Netherlands) |
Tutor, assisting students in the practical lessons |
|
|
Data Management for Quantitative Biology, University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
Tutor, managed seminar, hold tutorials, exam assistant |
NA |
Mentoring
| Time | University | Thesis | Links |
|---|---|---|---|
|
Master’s degree in Bioinformatics, University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
Joining medical data and pangenome graphs using the semantic web |
|
|
Bachelor’s degree in Bioinformatics, University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
Die Konstruktion eines Lodderomyces elongisporus Pangenomgraphen |
NA |
Courses
| Time | Place | Title | Links |
|---|---|---|---|
|
Comenius University in Bratislava, Faculty of Natural Sciences (wet lab), and Faculty of Mathematics, Physics and Informatics (analysis), Bratislava, Slovakia |
PANGAIA Winter Wet Lab School |
|
|
University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
Biomedical Data Science Symposium |
|
|
University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
Nextflow Workshop |
|
|
EMBO | EMBL Symposium: From Single- to Multiomics: Applications and Challenges in Data Integration |
||
|
Leibniz Institute of Plant Biochemistry, Halle (Saale) |
Joint CIBI user meeting 2017 on OpenMS, MetFrag and SeqAn |
|
|
University of Tübingen, Quantitative Biology Center (QBiC), Tübingen, Germany |
2nd Annual European Bioinformatics Core Community Workshop |
|
|
Vrije Universiteit, Amsterdam, Netherlands |
iGEM 2012 European Regional Jamboree |
Misc
| Time | Info | What | Links |
|---|---|---|---|
|
Reviewer for UK Research and Innovation |
||
|
Reviewer for Oxford Bioinformatics |
NA |
|
|
Associate Member of the Human Pangenome Reference Consortium |